Note
Go to the end to download the full example code.
Cache mechanism#
This example shows how EstimatorReport and
CrossValidationReport use caching to speed up computations.
Loading some data#
First, we load a dataset from skrub. Our goal is to predict if a company paid a
physician. The ultimate goal is to detect potential conflict of interest when it comes
to the actual problem that we want to solve.
from skrub.datasets import fetch_open_payments
dataset = fetch_open_payments()
df = dataset.X
y = dataset.y
from skrub import TableReport
TableReport(df)
| Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name | Dispute_Status_for_Publication | Name_of_Associated_Covered_Device_or_Medical_Supply1 | Name_of_Associated_Covered_Drug_or_Biological1 | Physician_Specialty | |
|---|---|---|---|---|---|
| 0 | ELI LILLY AND COMPANY | No | Allopathic & Osteopathic Physicians|Pediatrics|Pediatric Rheumatology | ||
| 1 | ELI LILLY AND COMPANY | No | Allopathic & Osteopathic Physicians|Internal Medicine|Nephrology | ||
| 2 | ELI LILLY AND COMPANY | No | Allopathic & Osteopathic Physicians|Internal Medicine|Rheumatology | ||
| 3 | ELI LILLY AND COMPANY | No | Allopathic & Osteopathic Physicians|Internal Medicine|Endocrinology, Diabetes & Metabolism | ||
| 4 | ELI LILLY AND COMPANY | No | EFFIENT | Allopathic & Osteopathic Physicians|Pediatrics|Pediatric Hematology-Oncology | |
| 73,553 | GlaxoSmithKline, LLC. | No | ZIAGEN | ||
| 73,554 | ALERE SCARBOROUGH, INC. | No | Alere PBP2a | ||
| 73,555 | NovoCure Limited | No | |||
| 73,556 | Wright Medical Technology, Inc. | No | HIPS | ||
| 73,557 | Alcon Research Ltd | No | Express |
Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name
ObjectDType- Null values
- 0 (0.0%)
- Unique values
-
1,466 (2.0%)
This column has a high cardinality (> 40).
Most frequent values
Merck Sharp & Dohme Corporation
Novartis Pharmaceuticals Corporation
Pfizer Inc.
Boston Scientific Corporation
Covidien Sales LLC
Stryker Corporation
SANOFI-AVENTIS U.S. LLC
AstraZeneca Pharmaceuticals LP
Genentech USA, Inc.
List:AbbVie, Inc.
['Merck Sharp & Dohme Corporation', 'Novartis Pharmaceuticals Corporation', 'Pfizer Inc.', 'Boston Scientific Corporation', 'Covidien Sales LLC', 'Stryker Corporation', 'SANOFI-AVENTIS U.S. LLC', 'AstraZeneca Pharmaceuticals LP', 'Genentech USA, Inc.', 'AbbVie, Inc.']
Dispute_Status_for_Publication
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (< 0.1%)
Most frequent values
No
Yes
['No', 'Yes']
Name_of_Associated_Covered_Device_or_Medical_Supply1
ObjectDType- Null values
- 43,088 (58.6%)
- Unique values
-
4,372 (5.9%)
This column has a high cardinality (> 40).
Most frequent values
Vascular
Spine
ARTHREX PRODUCT LINE DISTAL EXTREMITY ARTHROSCOPY
Surgical
ALL ARTHREX PRODUCT LINES
LifeVest
Spinal Cord Neurostimulation - Neuro
Da Vinci Surgical System
PAIN MANAGEMENT
List:Interventional Therapies
['Vascular', 'Spine', 'ARTHREX PRODUCT LINE DISTAL EXTREMITY ARTHROSCOPY', 'Surgical', 'ALL ARTHREX PRODUCT LINES', 'LifeVest', 'Spinal Cord Neurostimulation - Neuro', 'Da Vinci Surgical System', 'PAIN MANAGEMENT', 'Interventional Therapies']
Name_of_Associated_Covered_Drug_or_Biological1
ObjectDType- Null values
- 36,233 (49.3%)
- Unique values
-
2,262 (3.1%)
This column has a high cardinality (> 40).
Most frequent values
Invokana
Xarelto
NON-PRODUCT
Prolia
BUTRANS
NON BRAND
No Product
Nesina
ELIQUIS
List:Zytiga
['Invokana', 'Xarelto', 'NON-PRODUCT', 'Prolia', 'BUTRANS', 'NON BRAND', 'No Product', 'Nesina', 'ELIQUIS', 'Zytiga']
Physician_Specialty
ObjectDType- Null values
- 3,996 (5.4%)
- Unique values
-
513 (0.7%)
This column has a high cardinality (> 40).
Most frequent values
Allopathic & Osteopathic Physicians|Internal Medicine
Other Service Providers|Specialist
Allopathic & Osteopathic Physicians|Surgery
Allopathic & Osteopathic Physicians|Family Medicine
Allopathic & Osteopathic Physicians|Orthopaedic Surgery
Allopathic & Osteopathic Physicians|Internal Medicine|Cardiovascular Disease
Allopathic & Osteopathic Physicians|Pediatrics
Allopathic & Osteopathic Physicians|Radiology|Diagnostic Radiology
Allopathic & Osteopathic Physicians|Obstetrics & Gynecology
List:Student, Health Care|Student in an Organized Health Care Education/Training Program
['Allopathic & Osteopathic Physicians|Internal Medicine', 'Other Service Providers|Specialist', 'Allopathic & Osteopathic Physicians|Surgery', 'Allopathic & Osteopathic Physicians|Family Medicine', 'Allopathic & Osteopathic Physicians|Orthopaedic Surgery', 'Allopathic & Osteopathic Physicians|Internal Medicine|Cardiovascular Disease', 'Allopathic & Osteopathic Physicians|Pediatrics', 'Allopathic & Osteopathic Physicians|Radiology|Diagnostic Radiology', 'Allopathic & Osteopathic Physicians|Obstetrics & Gynecology', 'Student, Health Care|Student in an Organized Health Care Education/Training Program']
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
|
Column
|
Column name
|
dtype
|
Is sorted
|
Null values
|
Unique values
|
Mean
|
Std
|
Min
|
Median
|
Max
|
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name | ObjectDType | False | 0 (0.0%) | 1466 (2.0%) | |||||
| 1 | Dispute_Status_for_Publication | ObjectDType | False | 0 (0.0%) | 2 (< 0.1%) | |||||
| 2 | Name_of_Associated_Covered_Device_or_Medical_Supply1 | ObjectDType | False | 43088 (58.6%) | 4372 (5.9%) | |||||
| 3 | Name_of_Associated_Covered_Drug_or_Biological1 | ObjectDType | False | 36233 (49.3%) | 2262 (3.1%) | |||||
| 4 | Physician_Specialty | ObjectDType | False | 3996 (5.4%) | 513 (0.7%) |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name
ObjectDType- Null values
- 0 (0.0%)
- Unique values
-
1,466 (2.0%)
This column has a high cardinality (> 40).
Most frequent values
Merck Sharp & Dohme Corporation
Novartis Pharmaceuticals Corporation
Pfizer Inc.
Boston Scientific Corporation
Covidien Sales LLC
Stryker Corporation
SANOFI-AVENTIS U.S. LLC
AstraZeneca Pharmaceuticals LP
Genentech USA, Inc.
List:AbbVie, Inc.
['Merck Sharp & Dohme Corporation', 'Novartis Pharmaceuticals Corporation', 'Pfizer Inc.', 'Boston Scientific Corporation', 'Covidien Sales LLC', 'Stryker Corporation', 'SANOFI-AVENTIS U.S. LLC', 'AstraZeneca Pharmaceuticals LP', 'Genentech USA, Inc.', 'AbbVie, Inc.']
Dispute_Status_for_Publication
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (< 0.1%)
Most frequent values
No
Yes
['No', 'Yes']
Name_of_Associated_Covered_Device_or_Medical_Supply1
ObjectDType- Null values
- 43,088 (58.6%)
- Unique values
-
4,372 (5.9%)
This column has a high cardinality (> 40).
Most frequent values
Vascular
Spine
ARTHREX PRODUCT LINE DISTAL EXTREMITY ARTHROSCOPY
Surgical
ALL ARTHREX PRODUCT LINES
LifeVest
Spinal Cord Neurostimulation - Neuro
Da Vinci Surgical System
PAIN MANAGEMENT
List:Interventional Therapies
['Vascular', 'Spine', 'ARTHREX PRODUCT LINE DISTAL EXTREMITY ARTHROSCOPY', 'Surgical', 'ALL ARTHREX PRODUCT LINES', 'LifeVest', 'Spinal Cord Neurostimulation - Neuro', 'Da Vinci Surgical System', 'PAIN MANAGEMENT', 'Interventional Therapies']
Name_of_Associated_Covered_Drug_or_Biological1
ObjectDType- Null values
- 36,233 (49.3%)
- Unique values
-
2,262 (3.1%)
This column has a high cardinality (> 40).
Most frequent values
Invokana
Xarelto
NON-PRODUCT
Prolia
BUTRANS
NON BRAND
No Product
Nesina
ELIQUIS
List:Zytiga
['Invokana', 'Xarelto', 'NON-PRODUCT', 'Prolia', 'BUTRANS', 'NON BRAND', 'No Product', 'Nesina', 'ELIQUIS', 'Zytiga']
Physician_Specialty
ObjectDType- Null values
- 3,996 (5.4%)
- Unique values
-
513 (0.7%)
This column has a high cardinality (> 40).
Most frequent values
Allopathic & Osteopathic Physicians|Internal Medicine
Other Service Providers|Specialist
Allopathic & Osteopathic Physicians|Surgery
Allopathic & Osteopathic Physicians|Family Medicine
Allopathic & Osteopathic Physicians|Orthopaedic Surgery
Allopathic & Osteopathic Physicians|Internal Medicine|Cardiovascular Disease
Allopathic & Osteopathic Physicians|Pediatrics
Allopathic & Osteopathic Physicians|Radiology|Diagnostic Radiology
Allopathic & Osteopathic Physicians|Obstetrics & Gynecology
List:Student, Health Care|Student in an Organized Health Care Education/Training Program
['Allopathic & Osteopathic Physicians|Internal Medicine', 'Other Service Providers|Specialist', 'Allopathic & Osteopathic Physicians|Surgery', 'Allopathic & Osteopathic Physicians|Family Medicine', 'Allopathic & Osteopathic Physicians|Orthopaedic Surgery', 'Allopathic & Osteopathic Physicians|Internal Medicine|Cardiovascular Disease', 'Allopathic & Osteopathic Physicians|Pediatrics', 'Allopathic & Osteopathic Physicians|Radiology|Diagnostic Radiology', 'Allopathic & Osteopathic Physicians|Obstetrics & Gynecology', 'Student, Health Care|Student in an Organized Health Care Education/Training Program']
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V | Pearson's Correlation |
|---|---|---|---|
| Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name | Name_of_Associated_Covered_Drug_or_Biological1 | 0.295 | |
| Name_of_Associated_Covered_Device_or_Medical_Supply1 | Name_of_Associated_Covered_Drug_or_Biological1 | 0.258 | |
| Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name | Name_of_Associated_Covered_Device_or_Medical_Supply1 | 0.121 | |
| Name_of_Associated_Covered_Device_or_Medical_Supply1 | Physician_Specialty | 0.0880 | |
| Dispute_Status_for_Publication | Name_of_Associated_Covered_Drug_or_Biological1 | 0.0858 | |
| Dispute_Status_for_Publication | Physician_Specialty | 0.0715 | |
| Name_of_Associated_Covered_Drug_or_Biological1 | Physician_Specialty | 0.0594 | |
| Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name | Physician_Specialty | 0.0577 | |
| Applicable_Manufacturer_or_Applicable_GPO_Making_Payment_Name | Dispute_Status_for_Publication | 0.0565 | |
| Dispute_Status_for_Publication | Name_of_Associated_Covered_Device_or_Medical_Supply1 | 0.00934 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
import pandas as pd
TableReport(pd.DataFrame(y))
| status | |
|---|---|
| 0 | disallowed |
| 1 | disallowed |
| 2 | disallowed |
| 3 | disallowed |
| 4 | disallowed |
| 73,553 | allowed |
| 73,554 | allowed |
| 73,555 | allowed |
| 73,556 | allowed |
| 73,557 | allowed |
status
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (< 0.1%)
Most frequent values
disallowed
allowed
['disallowed', 'allowed']
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
|
Column
|
Column name
|
dtype
|
Is sorted
|
Null values
|
Unique values
|
Mean
|
Std
|
Min
|
Median
|
Max
|
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | status | ObjectDType | True | 0 (0.0%) | 2 (< 0.1%) |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
status
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (< 0.1%)
Most frequent values
disallowed
allowed
['disallowed', 'allowed']
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
The dataset has over 70,000 records with only categorical features. Some categories are not well defined.
Caching with EstimatorReport and CrossValidationReport#
We use skrub to create a simple predictive model that handles our dataset’s
challenges.
from skrub import tabular_learner
model = tabular_learner("classifier")
model
/opt/hostedtoolcache/Python/3.12.11/x64/lib/python3.12/site-packages/skrub/_tabular_pipeline.py:75: FutureWarning:
tabular_learner will be deprecated in the next release. Equivalent functionality is available in skrub.tabular_pipeline.
Pipeline(steps=[('tablevectorizer',
TableVectorizer(low_cardinality=ToCategorical())),
('histgradientboostingclassifier',
HistGradientBoostingClassifier())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Parameters
| steps | [('tablevectorizer', ...), ('histgradientboostingclassifier', ...)] | |
| transform_input | None | |
| memory | None | |
| verbose | False |
Parameters
| cardinality_threshold | 40 | |
| low_cardinality | ToCategorical() | |
| high_cardinality | StringEncoder() | |
| numeric | PassThrough() | |
| datetime | DatetimeEncoder() | |
| specific_transformers | () | |
| drop_null_fraction | 1.0 | |
| drop_if_constant | False | |
| drop_if_unique | False | |
| datetime_format | None | |
| n_jobs | None |
Parameters
Parameters
| resolution | 'hour' | |
| add_weekday | False | |
| add_total_seconds | True | |
| add_day_of_year | False | |
| periodic_encoding | None |
Parameters
Parameters
| n_components | 30 | |
| vectorizer | 'tfidf' | |
| ngram_range | (3, ...) | |
| analyzer | 'char_wb' | |
| stop_words | None | |
| random_state | None |
Parameters
| loss | 'log_loss' | |
| learning_rate | 0.1 | |
| max_iter | 100 | |
| max_leaf_nodes | 31 | |
| max_depth | None | |
| min_samples_leaf | 20 | |
| l2_regularization | 0.0 | |
| max_features | 1.0 | |
| max_bins | 255 | |
| categorical_features | 'from_dtype' | |
| monotonic_cst | None | |
| interaction_cst | None | |
| warm_start | False | |
| early_stopping | 'auto' | |
| scoring | 'loss' | |
| validation_fraction | 0.1 | |
| n_iter_no_change | 10 | |
| tol | 1e-07 | |
| verbose | 0 | |
| random_state | None | |
| class_weight | None |
This model handles all types of data: numbers, categories, dates, and missing values. Let’s train it on part of our dataset.
from skore import train_test_split
X_train, X_test, y_train, y_test = train_test_split(df, y, random_state=42)
# Let's keep a completely separate dataset
X_train, X_external, y_train, y_external = train_test_split(
X_train, y_train, random_state=42
)
╭───────────────────────────── HighClassImbalanceWarning ──────────────────────────────╮
│ It seems that you have a classification problem with a high class imbalance. In this │
│ case, using train_test_split may not be a good idea because of high variability in │
│ the scores obtained on the test set. To tackle this challenge we suggest to use │
│ skore's CrossValidationReport with the `splitter` parameter of your choice. │
╰──────────────────────────────────────────────────────────────────────────────────────╯
╭───────────────────────────────── ShuffleTrueWarning ─────────────────────────────────╮
│ We detected that the `shuffle` parameter is set to `True` either explicitly or from │
│ its default value. In case of time-ordered events (even if they are independent), │
│ this will result in inflated model performance evaluation because natural drift will │
│ not be taken into account. We recommend setting the shuffle parameter to `False` in │
│ order to ensure the evaluation process is really representative of your production │
│ release process. │
╰──────────────────────────────────────────────────────────────────────────────────────╯
╭───────────────────────────── HighClassImbalanceWarning ──────────────────────────────╮
│ It seems that you have a classification problem with a high class imbalance. In this │
│ case, using train_test_split may not be a good idea because of high variability in │
│ the scores obtained on the test set. To tackle this challenge we suggest to use │
│ skore's CrossValidationReport with the `splitter` parameter of your choice. │
╰──────────────────────────────────────────────────────────────────────────────────────╯
╭───────────────────────────────── ShuffleTrueWarning ─────────────────────────────────╮
│ We detected that the `shuffle` parameter is set to `True` either explicitly or from │
│ its default value. In case of time-ordered events (even if they are independent), │
│ this will result in inflated model performance evaluation because natural drift will │
│ not be taken into account. We recommend setting the shuffle parameter to `False` in │
│ order to ensure the evaluation process is really representative of your production │
│ release process. │
╰──────────────────────────────────────────────────────────────────────────────────────╯
Caching the predictions for fast metric computation#
First, we focus on EstimatorReport, as the same philosophy will
apply to CrossValidationReport.
Let’s explore how EstimatorReport uses caching to speed up
predictions. We start by training the model:
from skore import EstimatorReport
report = EstimatorReport(
model, X_train=X_train, y_train=y_train, X_test=X_test, y_test=y_test
)
report.help()
╭───────────── Tools to diagnose estimator HistGradientBoostingClassifier ─────────────╮
│ EstimatorReport │
│ ├── .metrics │
│ │ ├── .accuracy(...) (↗︎) - Compute the accuracy score. │
│ │ ├── .brier_score(...) (↘︎) - Compute the Brier score. │
│ │ ├── .confusion_matrix(...) - Plot the confusion matrix. │
│ │ ├── .log_loss(...) (↘︎) - Compute the log loss. │
│ │ ├── .precision(...) (↗︎) - Compute the precision score. │
│ │ ├── .precision_recall(...) - Plot the precision-recall curve. │
│ │ ├── .recall(...) (↗︎) - Compute the recall score. │
│ │ ├── .roc(...) - Plot the ROC curve. │
│ │ ├── .roc_auc(...) (↗︎) - Compute the ROC AUC score. │
│ │ ├── .timings(...) - Get all measured processing times related │
│ │ │ to the estimator. │
│ │ ├── .custom_metric(...) - Compute a custom metric. │
│ │ └── .summarize(...) - Report a set of metrics for our estimator. │
│ ├── .feature_importance │
│ │ └── .permutation(...) - Report the permutation feature importance. │
│ ├── .data │
│ │ └── .analyze(...) - Plot dataset statistics. │
│ ├── .cache_predictions(...) - Cache estimator's predictions. │
│ ├── .clear_cache(...) - Clear the cache. │
│ ├── .get_predictions(...) - Get estimator's predictions. │
│ └── Attributes │
│ ├── .X_test - Testing data │
│ ├── .X_train - Training data │
│ ├── .y_test - Testing target │
│ ├── .y_train - Training target │
│ ├── .estimator - Estimator to make the report from │
│ ├── .estimator_ - The cloned or copied estimator │
│ ├── .estimator_name_ - The name of the estimator │
│ ├── .fit - Whether to fit the estimator on the │
│ │ training data │
│ ├── .fit_time_ - The time taken to fit the estimator, in │
│ │ seconds │
│ ├── .ml_task - No description available │
│ └── .pos_label - For binary classification, the positive │
│ class │
│ │
│ │
│ Legend: │
│ (↗︎) higher is better (↘︎) lower is better │
╰──────────────────────────────────────────────────────────────────────────────────────╯
We compute the accuracy on our test set and measure how long it takes:
0.9514953779227842
Time taken: 1.61 seconds
For comparison, here’s how scikit-learn computes the same accuracy score:
from sklearn.metrics import accuracy_score
start = time.time()
result = accuracy_score(report.y_test, report.estimator_.predict(report.X_test))
end = time.time()
result
0.9514953779227842
Time taken: 1.59 seconds
Both approaches take similar time.
Now, watch what happens when we compute the accuracy again with our skore estimator report:
0.9514953779227842
Time taken: 0.00 seconds
The second calculation is instant! This happens because the report saves previous calculations in its cache. Let’s look inside the cache:
{(np.int64(6536099849484543332), None, 'predict', 'test', None): array(['disallowed', 'disallowed', 'disallowed', ..., 'disallowed',
'disallowed', 'disallowed'], shape=(18390,), dtype=object), (np.int64(6536099849484543332), 'test', None, 'predict_time'): 1.5970114840000065, (np.int64(6536099849484543332), 'accuracy_score', 'test'): 0.9514953779227842}
The cache stores predictions by type and data source. This means that computing metrics that use the same type of predictions will be faster. Let’s try the precision metric:
{'allowed': 0.661520190023753, 'disallowed': 0.9654091634374288}
Time taken: 0.06 seconds
We observe that it takes only a few milliseconds to compute the precision because we don’t need to re-compute the predictions and only have to compute the precision metric itself. Since the predictions are the bottleneck in terms of computation time, we observe an interesting speedup.
Caching all the possible predictions at once#
We can pre-compute all predictions at once using parallel processing:
report.cache_predictions(n_jobs=4)
Now, all possible predictions are stored. Any metric calculation will be much faster, even on different data (like the training set):
0.09382909512179063
Time taken: 0.09 seconds
Caching external data#
The report can also work with external data. We use data_source="X_y" to indicate
that we want to pass those external data.
start = time.time()
result = report.metrics.log_loss(data_source="X_y", X=X_external, y=y_external)
end = time.time()
result
0.12748818888907942
Time taken: 1.36 seconds
The first calculation of the above cell is slower than when using the internal train or test sets because it needs to compute a hash of the new data for later retrieval. Let’s calculate it again:
start = time.time()
result = report.metrics.log_loss(data_source="X_y", X=X_external, y=y_external)
end = time.time()
result
0.12748818888907942
Time taken: 0.14 seconds
It is much faster for the second time as the predictions are cached! The remaining time corresponds to the hash computation. Let’s compute the ROC AUC on the same data:
start = time.time()
result = report.metrics.roc_auc(data_source="X_y", X=X_external, y=y_external)
end = time.time()
result
0.933101925028547
Time taken: 0.16 seconds
We observe that the computation is already efficient because it boils down to two computations: the hash of the data and the ROC-AUC metric. We save a lot of time because we don’t need to re-compute the predictions.
Caching for plotting#
The cache also speeds up plots. Let’s create a ROC curve:
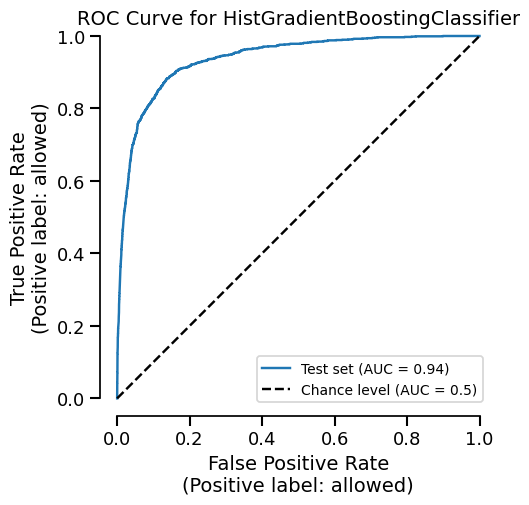
Time taken: 0.03 seconds
The second plot is instant because it uses cached data:
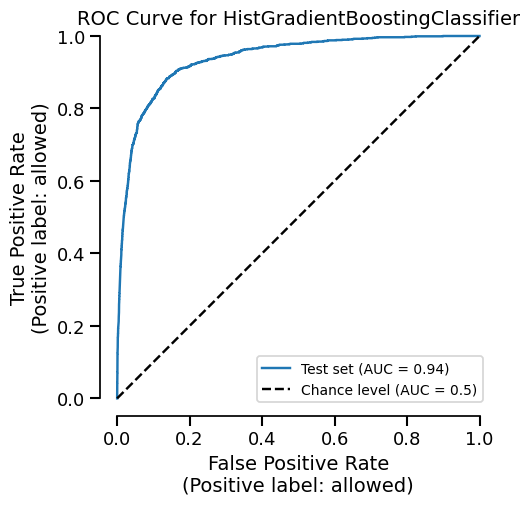
Time taken: 0.01 seconds
We only use the cache to retrieve the display object and not directly the matplotlib
figure. It means that we can still customize the cached plot before displaying it:
display.plot(roc_curve_kwargs={"color": "tab:orange"})
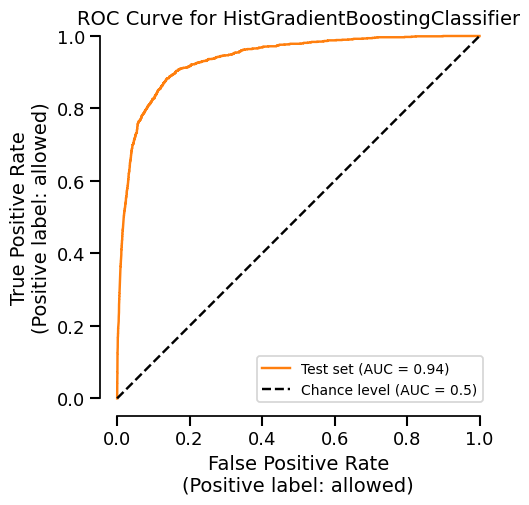
Be aware that we can clear the cache if we want to:
{}
It means that nothing is stored anymore in the cache.
Caching with CrossValidationReport#
CrossValidationReport uses the same caching system for each split
in cross-validation by leveraging the previous EstimatorReport:
from skore import CrossValidationReport
report = CrossValidationReport(model, X=df, y=y, splitter=5, n_jobs=4)
report.help()
╭───────────── Tools to diagnose estimator HistGradientBoostingClassifier ─────────────╮
│ CrossValidationReport │
│ ├── .metrics │
│ │ ├── .accuracy(...) (↗︎) - Compute the accuracy score. │
│ │ ├── .brier_score(...) (↘︎) - Compute the Brier score. │
│ │ ├── .log_loss(...) (↘︎) - Compute the log loss. │
│ │ ├── .precision(...) (↗︎) - Compute the precision score. │
│ │ ├── .precision_recall(...) - Plot the precision-recall curve. │
│ │ ├── .recall(...) (↗︎) - Compute the recall score. │
│ │ ├── .roc(...) - Plot the ROC curve. │
│ │ ├── .roc_auc(...) (↗︎) - Compute the ROC AUC score. │
│ │ ├── .timings(...) - Get all measured processing times related │
│ │ │ to the estimator. │
│ │ ├── .custom_metric(...) - Compute a custom metric. │
│ │ └── .summarize(...) - Report a set of metrics for our estimator. │
│ ├── .cache_predictions(...) - Cache the predictions for sub-estimators │
│ │ reports. │
│ ├── .clear_cache(...) - Clear the cache. │
│ ├── .get_predictions(...) - Get estimator's predictions. │
│ └── Attributes │
│ ├── .X - The data to fit │
│ ├── .y - The target variable to try to predict in │
│ │ the case of supervised learning │
│ ├── .estimator - Estimator to make the cross-validation │
│ │ report from │
│ ├── .estimator_ - The cloned or copied estimator │
│ ├── .estimator_name_ - The name of the estimator │
│ ├── .estimator_reports_ - The estimator reports for each split │
│ ├── .ml_task - No description available │
│ ├── .n_jobs - Number of jobs to run in parallel │
│ ├── .pos_label - For binary classification, the positive │
│ │ class │
│ ├── .split_indices - No description available │
│ └── .splitter - Determines the cross-validation splitting │
│ strategy │
│ │
│ │
│ Legend: │
│ (↗︎) higher is better (↘︎) lower is better │
╰──────────────────────────────────────────────────────────────────────────────────────╯
Since a CrossValidationReport uses many
EstimatorReport, we will observe the same behaviour as we previously
exposed.
The first call will be slow because it computes the predictions for each split.
| HistGradientBoostingClassifier | |||
|---|---|---|---|
| mean | std | ||
| Metric | Label / Average | ||
| Precision | allowed | 0.432482 | 0.123726 |
| disallowed | 0.959970 | 0.005586 | |
| Recall | allowed | 0.423435 | 0.097387 |
| disallowed | 0.951590 | 0.043077 | |
| ROC AUC | 0.874609 | 0.030393 | |
| Brier score | 0.063774 | 0.033356 | |
| Fit time (s) | 16.128872 | 3.197235 | |
| Predict time (s) | 2.083737 | 0.450096 | |
Time taken: 11.60 seconds
But the subsequent calls are fast because the predictions are cached.
| HistGradientBoostingClassifier | |||
|---|---|---|---|
| mean | std | ||
| Metric | Label / Average | ||
| Precision | allowed | 0.432482 | 0.123726 |
| disallowed | 0.959970 | 0.005586 | |
| Recall | allowed | 0.423435 | 0.097387 |
| disallowed | 0.951590 | 0.043077 | |
| ROC AUC | 0.874609 | 0.030393 | |
| Brier score | 0.063774 | 0.033356 | |
| Fit time (s) | 16.128872 | 3.197235 | |
| Predict time (s) | 2.083737 | 0.450096 | |
Time taken: 0.00 seconds
Hence, we observe the same type of behaviour as we previously exposed.
Total running time of the script: (1 minutes 14.669 seconds)